TLDR; Pickle is slow, cPickle is faster, JSON is surprisingly fast, Marshal is fastest, but when using PyPy all fall before a humble text file parser.
I’ve been working on the Yandex Personalized Web Search challenge on kaggle, which requires me to read in a large amount of data stored as multi-line, tab-separated, records. The data is ragged, each record has sub-fields of variable length, and there are 34.5 million of them. There are too many records to load in memory, and starting out, I’m not quite sure what features will end up being important. To reduce the bottle-neck of reading the files from disk, I wanted to pre-process the data, and store it in some format which afforded me faster reading and writing off the disk, without going to a full blown database solution. I ended up testing the following formats:
- pickle (using both the pickle and cPickle modules), a module which can serialize just about any datas structure, but is only available for python. I’ve used this extensively to persistify derived data for work.
- JSON, which only supports a basic set of data-types (fine for this use case), but can be handled by a larger number of languages (irrelevant for this use case).
- Marshal, which is not really meant for general serialization, but I was curious about it’s internals. This is not really a recommended format, the documentation clearly warns “Details of the format are undocumented on purpose; it may change between Python versions.”
The speed test consisted of writing 50,000 records to disk, using each of the three formats above. Two approaches are taken to writing: A) For each record, call the dump method of the module. This method takes as input the object to serialize and a file handler, and appends the record to the file on disk:
for r in records: module.dump(r, ofile)
B) To reduce the number of file access calls, join the string representation of several records, and write them all to disk at once. This method is referred to as “bulk” writing in the plots below (N.B. for JSON, I join on ‘\n’, not the empty string ”):
ofile.write(''.join(module.dumps(r) for r in records))
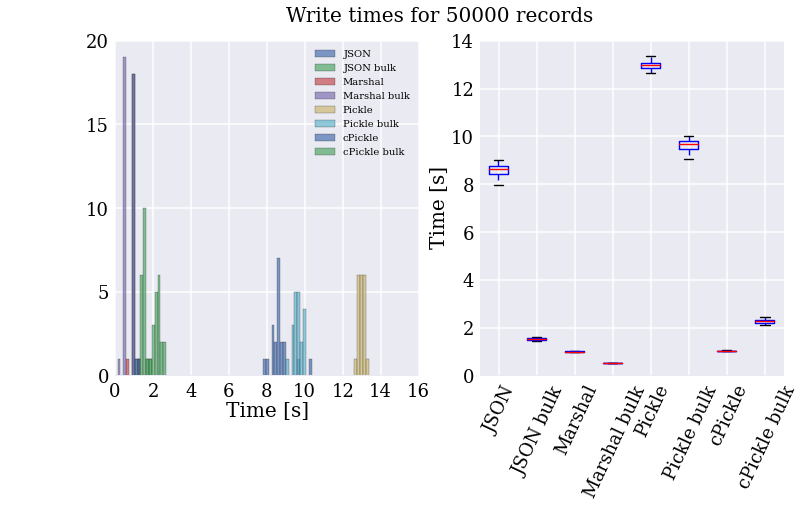
Here are the speed test results for writing. The histogram on the left shows the time per test, which is nicely summarized on the right for those who prefer whisker plots.
The resulting file sizes are all pretty-much in the same ball-park, with JSON a bit bigger (the cost of being fully human readable, I suppose).
- JSON: 39M
- Marshal: 27M
- Pickle/cPickle: 24M
A couple of interesting features pop out in the above plot and numbers:
- Marshal is over twice as fast as the next fastest method, and two orders of magnitude faster than the slowest method. This seems to be because it does less formatting of the data before serializing it.
- In most cases, “bulk” writing, where the string representation of multiple records is generated in one step, is faster than writing individual records. The only exception is using the cPickle module, where this is reversed. I have no idea why cPickle is different, it doesn’t make much sense intuitively, and couldn’t find any good explanations online.
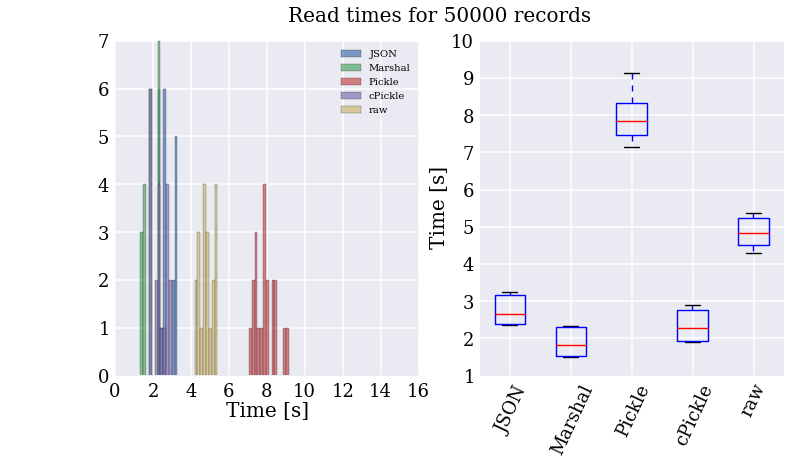
So, Marshal seems to be the fastest at writing the data to disk, but this is important only once when I pass through all the records to convert them. How do they compare when I perform a similar speed test to read in the records from the files generated in the previous step?
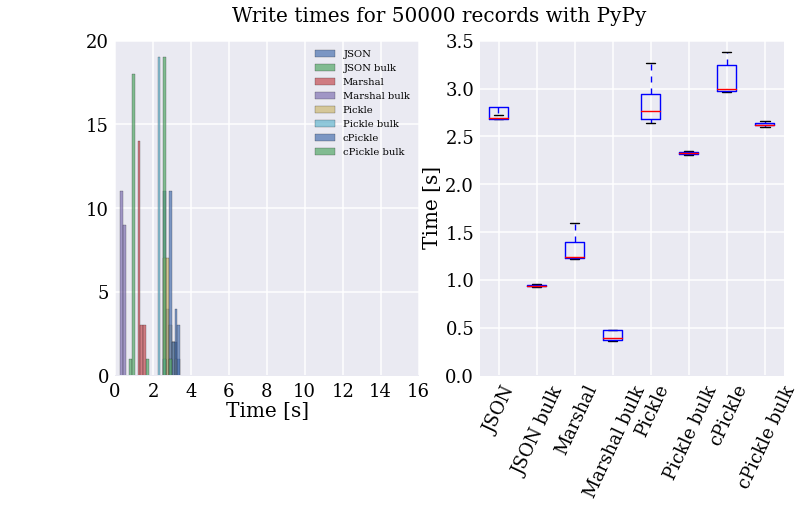
Well, looks like I’ll be using Marshal then, huh? Not so fast! This is all well and good, but much of the slowness of the text file parser comes from the for loop. Damn these interepreted languages and their inefficient byte-code. How do things change if we let the PyPy’s JIT compiler turn it into cached machine code?
My God! It’s full of stars!
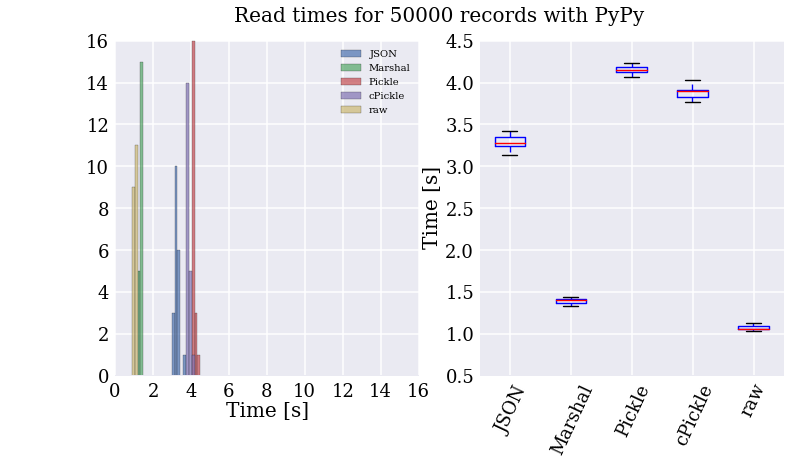
Let me just say that I am super impressed with PyPy (also, apparently, I wasted my time with writing up the code for all these formats). Reading the “raw” text file is now the fastest method of accessing the records, taking on average one second to read 50k records (about 150k lines of data with 4 to 26 fields of numbers).
I got a couple more speed-ups by optimizing the parser, the biggest of which came from using named tuples instead of dicts. Since these make use of the new-style classes’ “__slots__” attribute to remove the overhead of dynamic dict lookups. Bottom line, using PyPy and those additional modifications, I can read through all 34.5M records in under 20 minutes! Sampling from the records, I can prototype new features in a few minutes, which makes development much smoother.